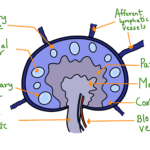
For most Molecular biology studies, it is crucial to quantify the product obtained after a PCR. Many methods have been discovered for this quantification, characterized under two categories: homogenous and heterogeneous methods. Heterogenous methods involve the separation of products from the PCR reagents and quantifying them. Methods like agarose gel electrophoresis and Ethidium Bromide (EtBr) to visualize the product fall under this category. In Homogenous methods, specific detection of amplicons is done in the reaction mixture itself without any separation or wash step. Many kinds of homogenous detection methods have been designed. Some of these methods make use of the chromogenic approaches, but the majority of the methods depend on the product’s fluorometric quantification. The different fluorometric quantification methods developed include using SYBR green dye, using TaqMan probes, using hybridization probes, and using melting probes. Let us discuss them one by one in this article.
Using SYBR green dye:
SYBR green dye is a unique kind of fluorescent dye that binds to the double-stranded DNA only. During the start of the PCR, the reaction mixture contains denatured DNA primers and dye. The unbound molecules of the dye fluoresce weakly, producing a weak signal which can be subtracted from the actual signal. Once the primer binds to the targeted part of the DNA and elongation is started, the SYBR green molecules bind to the double-stranded DNA and give a fluorescence signal. The fluorescence signal is highly enhanced during the elongation phase. Fluorescence drops again during the next cycle in a denaturation step as DNA molecules become single-stranded and SYBR green molecules detach from them. Fluorescence measurement is performed at the end of each elongation cycle to measure the amount of double-stranded DNA in a PCR mixture. One disadvantage of this technique is that as SYBR green bonds to double-stranded DNA only, it can bind to primer dimer formed during PCR and may give a false-positive result.

Using hybridization probes:
The hybridization probes are designed in an amplicon specific manner. The donor probe is labelled at the 3′ end with fluorescein, and the acceptor probe is labelled at the 5′ end with LightCycler Red. These probes are added to a PCR mixture along with forward and reverse primers. These two labelled probes anneal with the amplified single-stranded DNA fragment in a head-to-tail conformation in the annealing phase. The light from the LED of a machine excites the fluorescein, which emits green light after excitation. LightCycler Red then absorbs this green light in the form of energy. LightCycler Red becomes excited and emits a signal in the form of red light. This phenomenon is known as Forster Resonance Energy Transfer (FRET). The red fluorescence is measured at the end of each annealing step when fluorescence is highest. After the annealing step, the temperature is raised for the elongation step, where the DNA becomes double-stranded. The hybridization probes are displaced and are now too far apart for the FRET to take place.

Using TaqMan probes:
These types of probes are also designed to bind to the internal amplicon. In Taqman or Beacon probes, a fluorochrome (Molecule capable of generating fluorescence) and a quencher (Molecule responsible for the quenching of the fluorescence) is located on a single probe. The proximity between fluorophore and quencher is responsible for the suppression of the fluorescence of the fluorophore. Fluorescence is observed once this proximity is broken. In PCR, during the annealing step, TaqMan or Beacon probe binds internally to the target sequence. Fluorescence is minimal in this step as the quencher is still in the proximity of the fluorophore. In the extension step of the cycle, Taq polymerase extends the primer to form a double-stranded product. 5′ to 3′ exonuclease activity of the Taq polymerase breaks the proximity of fluorophore and quencher by making reporter free, which results in fluorescence. This fluorescence is detected at the end of the extension step of each cycle.