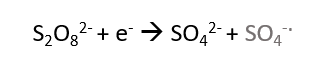What can be the simple technique to find out molecular weight of the in the given sample? Or what if we want to determine if the protein has a single subunit or multiple subunit?
The simple and most commonly used technique to answer the above questions is SDS-PAGE.
SDS-PAGE stands for Sodium Dodecyl Sulphate Polyacrylamide Gel Electrophoresis.
Sample preparation
The protein sample is pre-treated with anionic detergent-SDS, which denatures the protein by separating it into its protein subunits and then coating the negative charge over separated subunits, irrespective of their native charge (approximately, each SDS molecule associates with every two amino acid molecules). Also, another chemical, β-Mercaptoethanol is added to break the disulfide bond in the native state of the protein. Thus, in a sample, all the proteins have the same mass to charge ratio and shape. Thus, the only difference for separation is size or molecular weight of the protein.
Gel formation
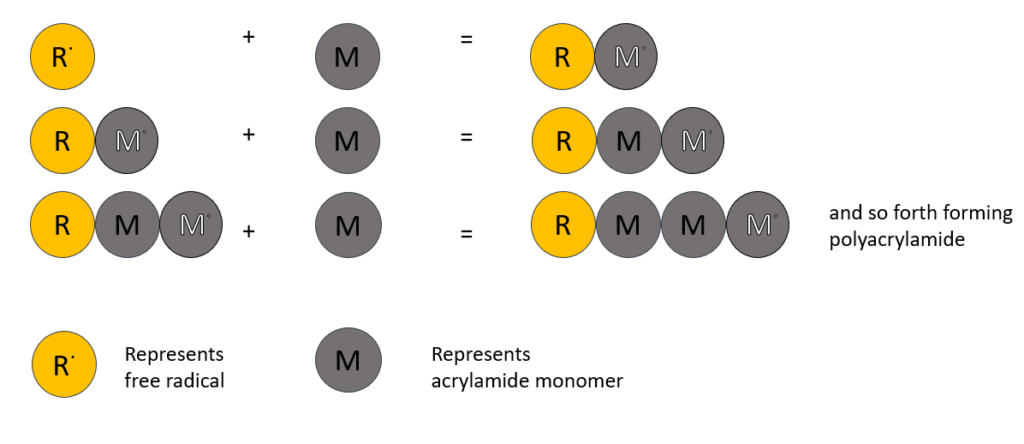
The acrylamide is an inert material used as a matrix whose porosity will be maintained by the amount of acrylamide concentration present. Acrylamide monomers are polymerized using a crosslinking agent, bis-acrylamide (it is basically two acrylamide molecules linked by a methylene group) into a long growing chain. The polymerization follows free-radical catalysis which is initiated by addition of ammonium persulphate (APS) and the base N,N,N’,N’-tetramethylenediamine (TEMED). The free radical is produced by decomposition of the persulphate ion catalyzed by TEMED.
Protein migration
The SDS-PAGE is a discontinuous gel system consisting of stacking gel with 6.8 pH and Separating gel with 8.8 pH. There is also a migration buffer with Tris-Glycine of pH 8.3, where the protein migrates from negative electrode to positive electrode. Glycine being a zwitterion ion helps in slow stacking since at lower pH (6.8 pH) it is acidic where its mobility decreases.
The stacking layer with large pore size has wells where protein samples are loaded, containing glycine, protein-SDS complex and chloride ions (Cl–), which will migrate in the same order from negative to positive electrode. Once it reaches the separating gel the pH changes to alkaline (8.8 pH) with smaller pore size. Here, the charge on glycine becomes negative thus order of migration changes to protein-SDS complex, glycine and chloride ions from negative to positive electrode. Thus, according to the size and molecular weight, the larger protein complex cannot migrate more towards the positive electrode due to smaller pore size, while smaller proteins migrate downward towards the positive electrode, hence separating proteins according to the molecular size!!
Molecular Weight Determination
To determine the molecular weight of an unknown protein the relative mobilities ( Rf ) of standard proteins loaded in the form of ladder on gel are determined and a graph is plotted for log molecular weight vs Rf values.
Rf = distance migrated by protein/ distance migrated by dye
Thus, Rf of unknown protein is determined and then the log Molecular weight can be calculated from the graph.