Each organism genome contains 1000s of genes, each gene (or DNA) is transcribed into RNA, which is eventually translated into protein. Blotting technique for the detection of RNA is known as Northern blotting. To study the level of gene expression and amount of RNA transcribed from each tissue, we can use the Northern Blotting Technique. We can study the cellular control over structure and function by determining the particular gene expression rates during differentiation, morphogenesis and abnormal or diseased conditions. The technique is used for detection of specific RNA sequences in total RNA extract. It was developed by James Alwine, David Kemp and George Stark at Stanford University (1977).
Principle
Northern blotting is a hybridization-based technique, in which RNA is separated on the basis of size (molecular weight) and detected on a nylon membrane using hybridization probe (radioactive labelled or chemiluminescence) with a base sequence complementary to all, or a part, of the sequence of the target mRNA.
- Electrophoresis: Separates RNA on the basis of their molecular weight and type in agarose gel having ethidium bromide (EtBr), an intercalating agent and using formaldehyde as the denaturing agent for RNA . The small size RNA, MicroRNA or fragmented RNA can be separated on denaturing polyacrylamide gel and here RNA is denatured using urea.
- Capillary action: The RNA bands move towards the membrane by capillary movement and entrap in sheet and buffer moves ahead.
- Hybridization: In molecular biology, it means the process of forming a double stranded nucleic acid from joining two complementary strands of DNA or RNA.
Procedure
RNA isolation from the sample
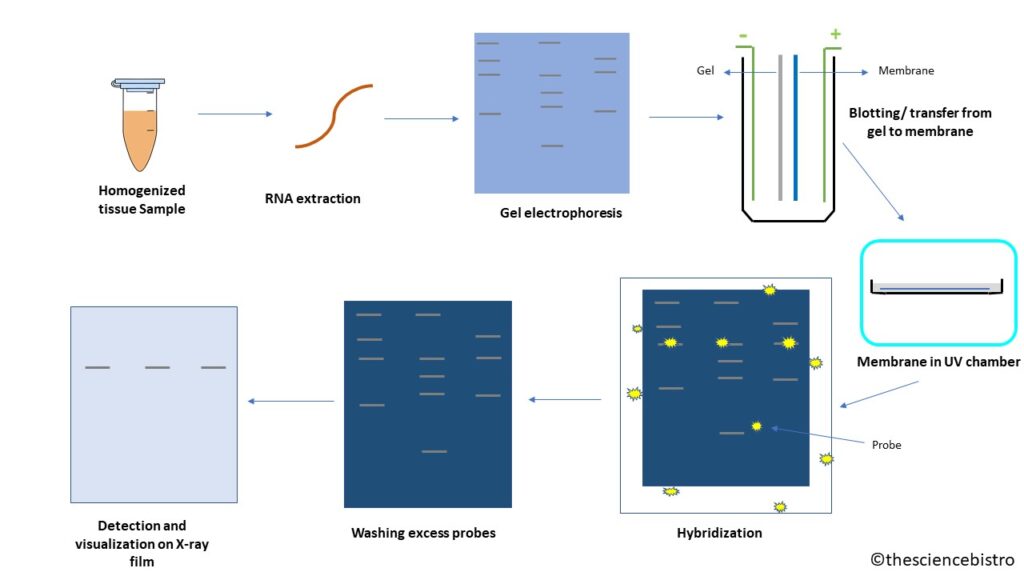
The process of Northern blotting starts with isolation of total RNA of the cells of homogenized tissue sample. It can have many types of RNA like transfer RNA(t-RNA), ribosomal RNA (rRNA), messenger RNA (mRNA), fragmented RNA and microRNA. For gene expression we check the levels of mRNA. The mRNAs are the only RNAs with poly(A) tail and using this characteristic it can be isolated using oligo (dT) cellulose chromatography.
Gel electrophoresis
The isolated sample are then separated by gel electrophoresis. Usually RNA is separated on agarose gel having a denaturing agent like formaldehyde. The agarose gel is stained with ethidium bromide (EtBr) and observed under UV light to know the quality and quantity of RNA. If the size of RNA is small like fragmented RNA or microRNA, then we can use polyacrylamide gel electrophoresis to separate RNA. The polyacrylamide can be stained with Coomassie Brilliant Blue to observe the bands. Both the gels are loaded with the ladder in one of the wells, as in to know the size of the fragment.
Blotting
In this process, the RNA is transferred from gel to the nylon membrane, since the gel being fragile, probes cannot enter the gel matrix. An alternative to nylon membrane is diazobenzyloxymethyl paper, which binds to RNA but its case specific (only when nitrocellulose or nylon does not work) The negatively charged RNA is transferred to positively charged nylon membrane via capillary action. the transfer takes place in buffer and a sandwich is made in sequence such hat sponge having gel, them membrane, plastic towels and weight over it in the following sequence. The bubbles are removed and current flows from negative electrode to positive, i.e., gel to membrane. Once the bands are transferred to the membrane, we can put the membrane in UV chamber/incubator. Here, the membrane would cross-link with the RNA present on it, so it is permanently fixed on nylon membrane.
Hybridization and washing
To complement RNA sequence of interest, the probe should be composed of nucleic acid like DNA, RNA or oligonucleotides. Usually cDNA is made with labelled primers for RNA of interest and this cDNA acts as a probe for northern blotting. The labelling of probes can be done either by radioactive isotopes (32P) or chemiluminescence. The radioactive probes being health hazardous are generally avoided, while chemiluminescence can be due to breakdown of chemiluminescent substrate due to alkaline phosphatase or horseradish peroxidase (HRP). The chemiluminescent labelling can occur in two ways:
- Probe is directly attached to the enzyme
- Probe is attached to a ligand and ligand attached to the enzyme.
After the hybridization of RNA of interest and probe is done, the excess probes must be washed away, to reduce the background noise.
Visualization
The detection of both radioactive labelled and chemiluminescent labelled sequences is done on X-ray film.
The chemiluminescent labelled probes are preferred over radioactive probes since they are more sensitive and faster and less health hazardous.